Welcome to Mao lab!
PUBLICATIONS
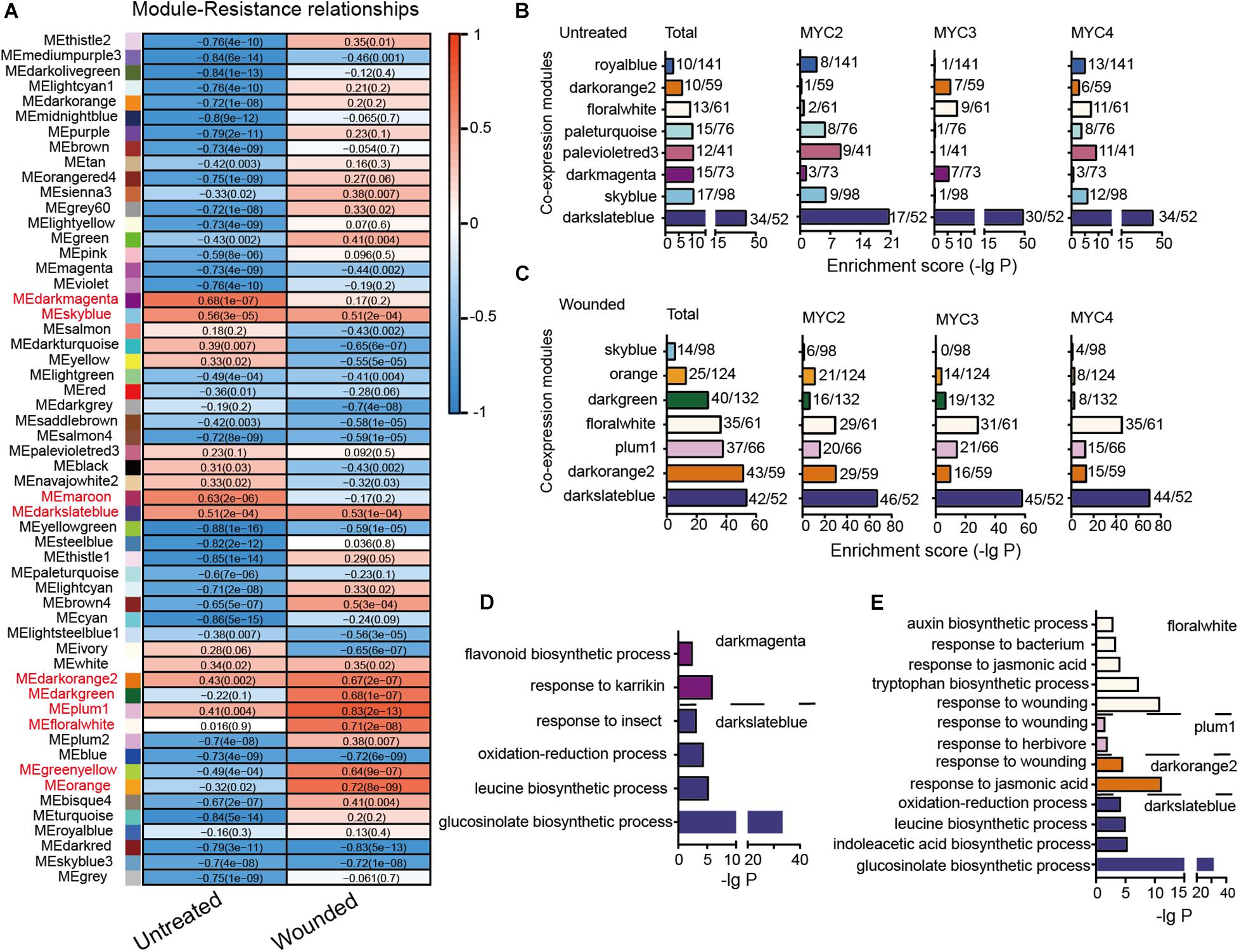
Differential Contributions of MYCs to Insect Defense Reveals Flavonoids Alleviating Growth Inhibition Caused by Wounding in Arabidopsis
Frontiers in plant science
2021
Abstract:
In Arabidopsis, basic helix–loop–helix transcription factors (TFs) MYC2, MYC3, and MYC4 are involved in many biological processes, such as defense against insects. We found that despite functional redundancy, MYC-related mutants displayed different resistance to cotton bollworm (Helicoverpa armigera). To screen out the most likely genes involved in defense against insects, we analyzed the correlation of gene expression with cotton bollworm resistance in wild-type (WT) and MYC-related mutants. In total, the expression of 94 genes in untreated plants and 545 genes in wounded plants were strongly correlated with insect resistance, and these genes were defined as MGAIs (MYC-related genes against insects). MYC3 had the greatest impact on the total expression of MGAIs. Gene ontology (GO) analysis revealed that besides the biosynthesis pathway of glucosinolates (GLSs), MGAIs, which are well-known defense compounds, were also enriched in flavonoid biosynthesis. Moreover, MYC3 dominantly affected the gene expression of flavonoid biosynthesis. Weighted gene co-expression network analysis (WGCNA) revealed that AAE18, which is involved in activating auxin precursor 2,4-dichlorophenoxybutyric acid (2,4-DB) and two other auxin response genes, was highly co-expressed with flavonoid biosynthesis genes. With wounding treatment, the WT plants exhibited better growth performance than chalcone synthase (CHS), which was defective in flavonoid biosynthesis. The data demonstrated dominant contributions of MYC3 to cotton bollworm resistance and imply that flavonoids might alleviate the growth inhibition caused by wounding in Arabidopsis.
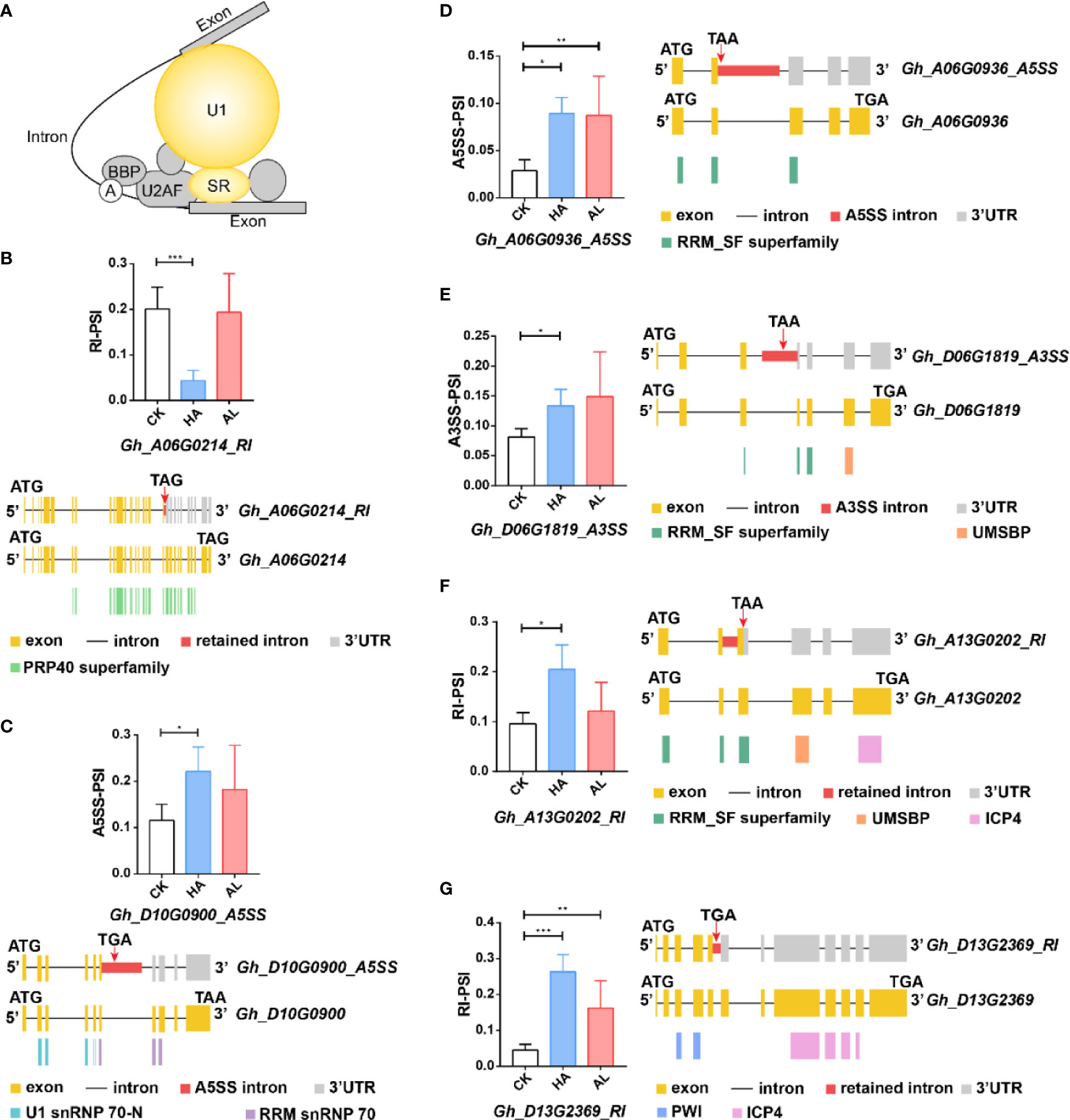
Differential transcription and alternative splicing in cotton underly specialized defense responses against pests
Frontiers in plant science
2020
Abstract:
The green mirid bug (Apolygus lucorum) and the cotton bollworm (Helicoverpa armigera) are both preferred to live on cotton but cause different symptoms, suggesting specialized responses of cotton to the two insects. In this study, we investigated differential molecular mechanisms underlying cotton plant defenses against A. lucorum and H. armigera via transcriptomic analyses. At the transcription level, jasmonate (JA) signaling was dominated in defense against H. armigera whereas salicylic acid (SA) signaling was more significant in defense against A. lucorum. A set of pathogenesis-related (PR) genes and protease inhibitor genes were differentially induced by the two insects. Insect infestations also had an impact on alternative splicing (AS), which was altered more significantly by the H. armigera than A. lucorum. Interestingly, most differential AS (DAS) genes had no obvious change at the transcription level. GO analysis revealed that biological process termed “RNA splicing” and “cellular response to abiotic stimulus” were enriched only in DAS genes from the H. armigera infested samples. Furthermore, insect infestations induced the retained intron of GhJAZs transcripts, which produced a truncated protein lacking the intact Jas motif. Taken together, our data demonstrate that the specialized cotton response to different insects is regulated by gene transcription and AS as well.

Research advances in plant–insect molecular interaction
F1000Research
2020
Abstract:
Acute and precise signal perception and transduction are essential for plant defense against insects. Insect elicitors—that is, the biologically active molecules from insects’ oral secretion (which contains regurgitant and saliva), frass, ovipositional fluids, and the endosymbionts—are recognized by plants and subsequently induce a local or systematic defense response. On the other hand, insects secrete various types of effectors to interfere with plant defense at multiple levels for better adaptation. Jasmonate is a main regulator involved in plant defense against insects and integrates with multiple pathways to make up the intricate defense network. Jasmonate signaling is strictly regulated in plants to avoid the hypersensitive defense response and seems to be vulnerable to assault by insect effectors at the same time. Here, we summarize recently identified elicitors, effectors, and their target proteins in plants and discuss their underlying molecular mechanisms.
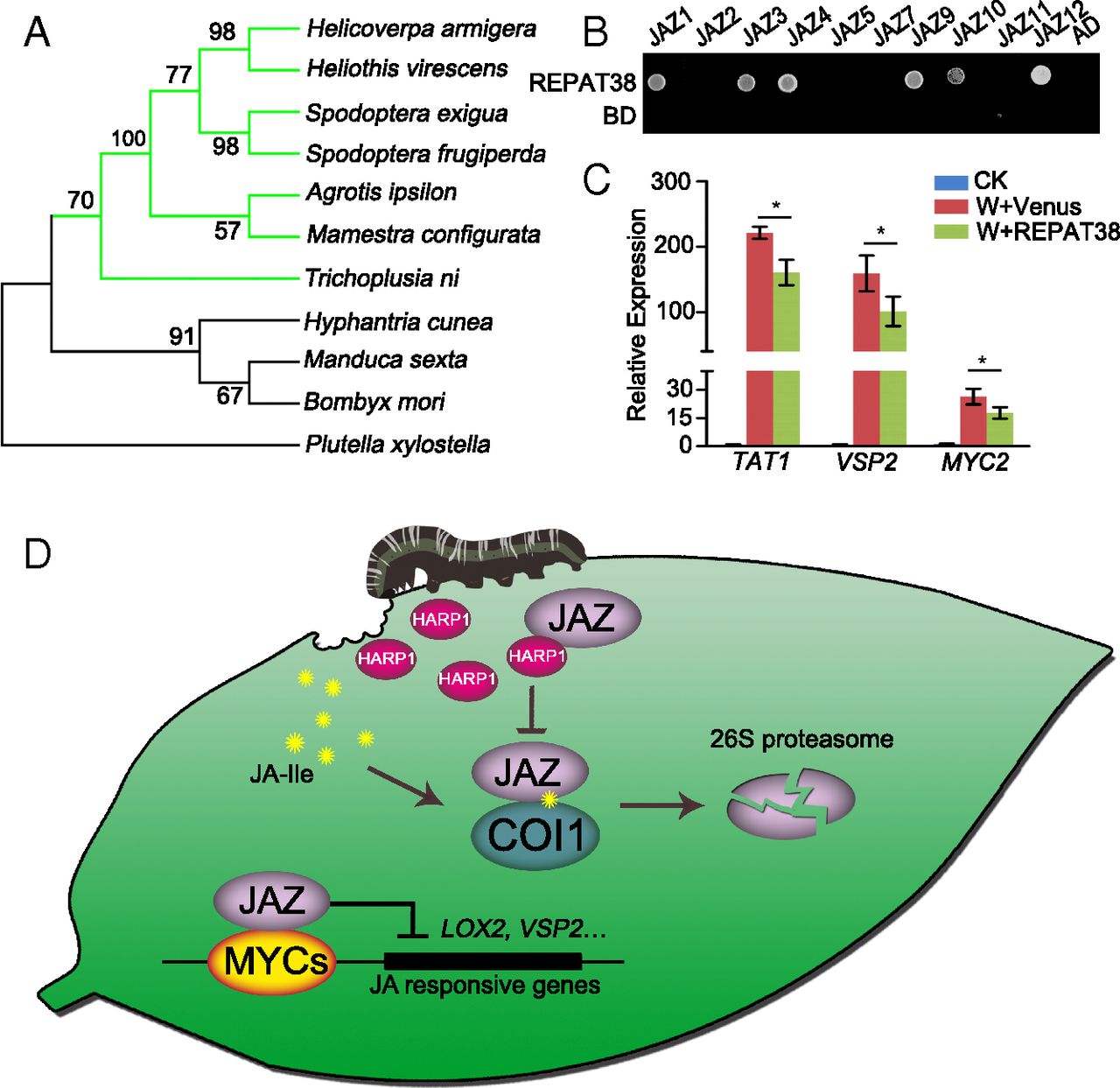
An effector from cotton bollworm oral secretion impairs host plant defense signaling
P.N.A.S.
2019
Abstract:
Insects have evolved effectors to conquer plant defense. Most known insect effectors are isolated from sucking insects, and examples from chewing insects are limited. Moreover, the targets of insect effectors in host plants remain unknown. Here, we address a chewing insect effector and its working mechanism. Cotton bollworm (Helicoverpa armigera) is a lepidopteran insect widely existing in nature and severely affecting crop productivity. We isolated an effector named HARP1 from H. armigera oral secretion (OS). HARP1 was released from larvae to plant leaves during feeding and entered into the plant cells through wounding sites. Expression of HARP1 in Arabidopsis mitigated the global expression of wounding and jasmonate (JA) responsive genes and rendered the plants more susceptible to insect feeding. HARP1 directly interacted with JASMONATE-ZIM-domain (JAZ) repressors to prevent the COI1-mediated JAZ degradation, thus blocking JA signaling transduction. HARP1-like proteins have conserved function as effectors in noctuidae, and these types of effectors might contribute to insect adaptation to host plants during coevolution.

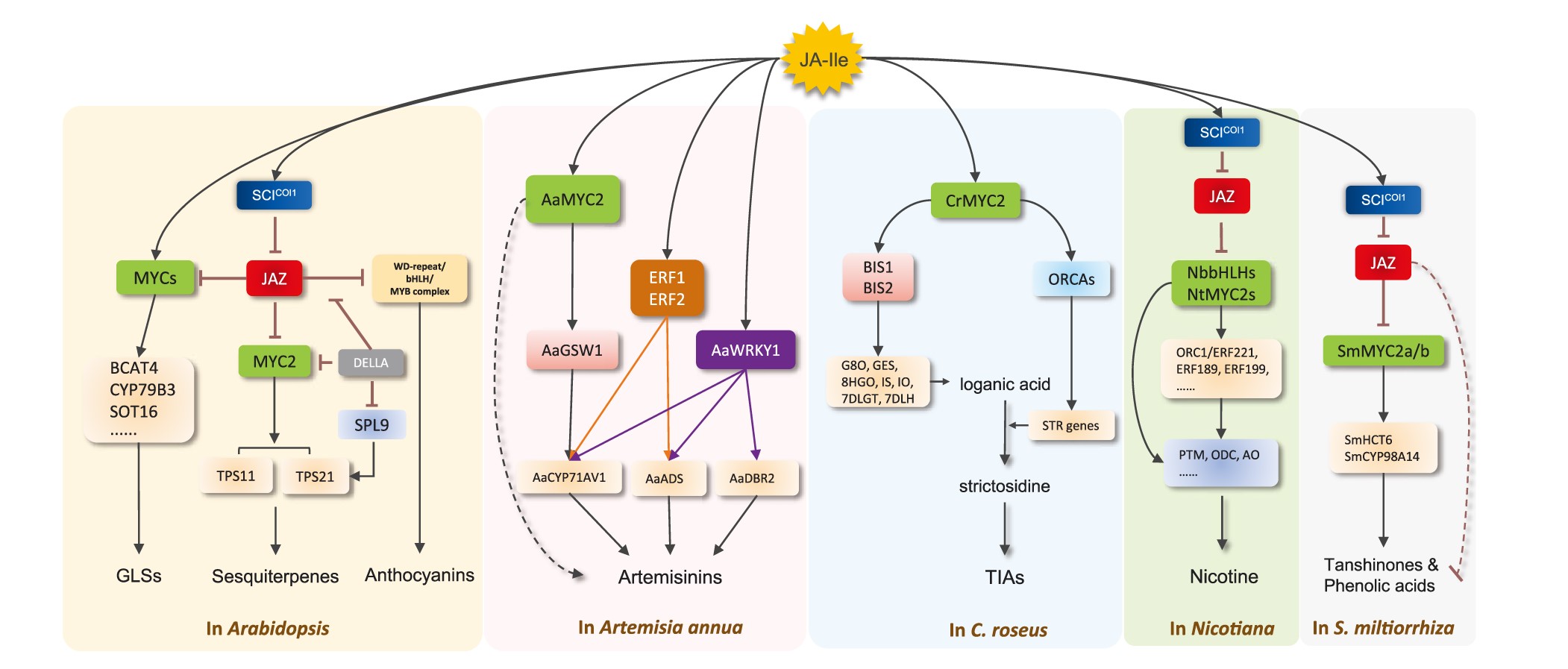
Plant Specialized Metabolism Regulated by Jasmonate Signaling
Plant and Cell Physiology
2019
Abstract:
As sessile and autotrophic organisms, plants have evolved sophisticated pathways to produce a rich array of specialized metabolites, many of which are biologically active and function as defense substances in protecting plants from herbivores and pathogens. Upon stimuli, these structurally diverse small molecules may be synthesized or constitutively accumulated. Jasmonate acids (JAs) are the major defense phytohormone involved in transducing external signals (such as wounding) to activate defense reactions, including, in particular, the reprogramming of metabolic pathways that initiate and enhance the production of defense compounds against insect herbivores and pathogens. In this review, we summarize the progress of recent research on the control of specialized metabolic pathways in plants by JA signaling, with an emphasis on the molecular regulation of terpene and alkaloid biosynthesis. We also discuss the interplay between JA signaling and various signaling pathways during plant defense responses. These studies provide valuable data for breeding insect-proof crops and pave the way to engineering the production of valuable metabolites in future.
Heterogeneous signals in plant–biotic interactions and their applications
Science China Life Sciences
2019
Abstract:
Plants have to overcome different types of environment stress including various insect and pathogen attacks during their life cycle. With long-term evolution, plants have developed sophisticated systems to recognize different biotic attacks and initiate an integrated defense network for survival. On the other hand, pathogens and insects have devised multiple strategies to adapt to their host plants. In the past three decades, substantial progress has been made toward improving the understanding of plant–biotic interactions. An increasing number of active molecules have been identified that not only have functions within an organism but also act as heterogeneous regulators that can migrate across interacting individuals (Kan et al., 2017; Qian et al., 2017). The discovery of new heterogeneous regulators and their mechanisms of migration across interacting organisms is potentially helpful in the development of novel strategies to protect crop plants from biotic threats.
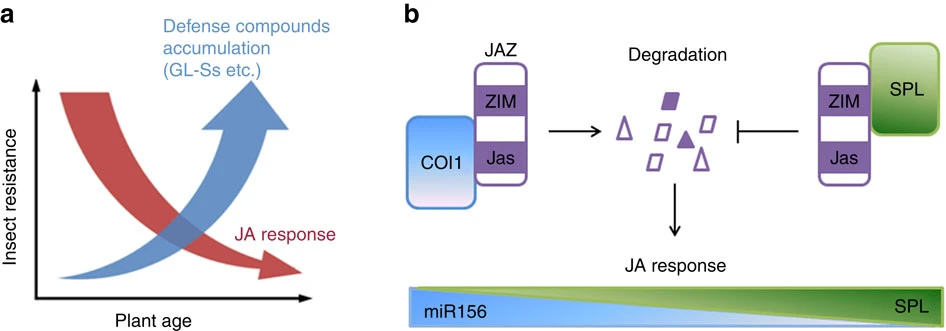
Jasmonate response decay and defense metabolite accumulation contributes to age-regulated dynamics of plant insect resistance
NATURE COMMUNICATIONS
2017
Abstract:
Immunity deteriorates with age in animals but comparatively little is known about the temporal regulation of plant resistance to herbivores. The phytohormone jasmonate (JA) is a key regulator of plant insect defense. Here, we show that the JA response decays progressively in Arabidopsis. We show that this decay is regulated by the miR156-targeted SQUAMOSA PROMOTER BINDING PROTEIN-LIKE9 (SPL9) group of proteins, which can interact with JA ZIM-domain (JAZ) proteins, including JAZ3. As SPL9 levels gradually increase, JAZ3 accumulates and the JA response is attenuated. We provide evidence that this pathway contributes to insect resistance in young plants. Interestingly however, despite the decay in JA response, older plants are still comparatively more resistant to both the lepidopteran generalist Helicoverpa armigera and the specialist Plutella xylostella, along with increased accumulation of glucosinolates. We propose a model whereby constitutive accumulation of defense compounds plays a role in compensating for age-related JA-response attenuation during plant maturation.
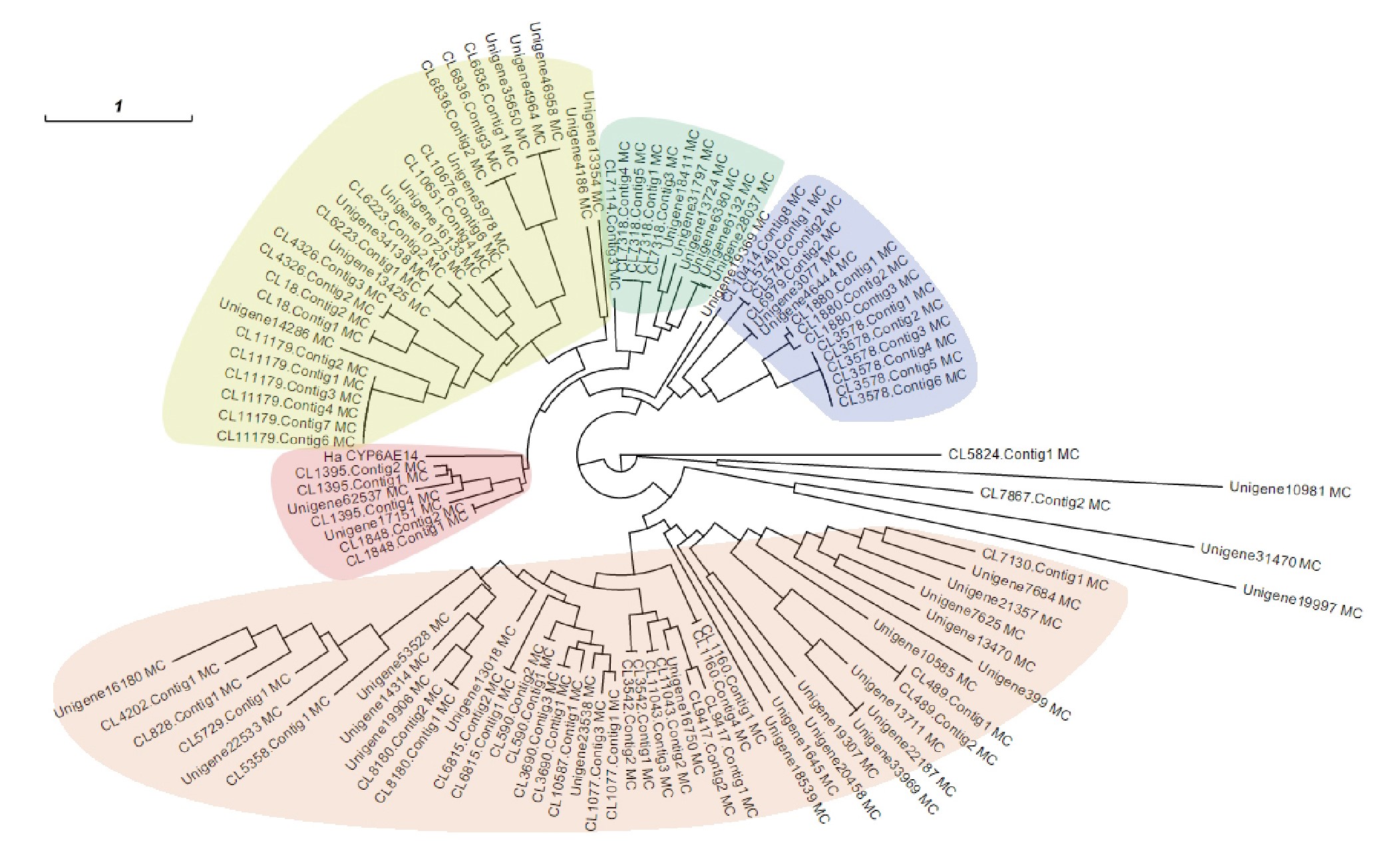
Transcriptome analysis of three cotton pests reveals features of gene expressions in the mesophyll feeder Apolygus lucorum
Science China Life Sciences
2017
Abstract:
The green mirid bug Apolygus lucorum is an agricultural pest that is known to cause damage to more than 150 plant species. Here, we report the transcriptomes of A. lucorum at three different developmental stages (the second and fifth instar nymphs and adults). A total of 98,236 unigenes with an average length of 1,335 nt was obtained, of which 50,640 were annotated, including those encoding digestive enzymes and cytochrome P450s. Comparisons with cotton bollworm and cotton aphid transcriptomes revealed distinct features of A. lucorum as a mesophyll feeder. The gene expression dynamics varied during development from young nymphs to adults. The high-quality transcriptome data and the gene expression dynamics reported here provide valuable data for a more comprehensive understanding of the physiology and development of mirid bugs, and for mining targets for their control.