Welcome to Mao lab!
An effector from cotton bollworm oral secretion impairs host plant defense signaling
Abstract Insects have evolved effectors to conquer plant defense. Most known insect effectors are isolated from sucking insects, and examples from chewing insects are limited. Moreover, the targets of insect effectors in host plants remain unknown. Here, we address a chewing insect effector and its working mechanism. Cotton bollworm (Helicoverpa armigera) is a lepidopteran insect widely existing in nature and severely affecting crop productivity. We isolated an effector named HARP1 from H. armigera oral secretion (OS). HARP1 was released from larvae to plant leaves during feeding and entered into the plant cells through wounding sites. Expression of HARP1 in Arabidopsis mitigated the global expression of wounding and jasmonate (JA) responsive genes and rendered the plants more susceptible to insect feeding. HARP1 directly interacted with JASMONATE-ZIM-domain (JAZ) repressors to prevent the COI1-mediated JAZ degradation, thus blocking JA signaling transduction. HARP1-like proteins have conserved function as effectors in noctuidae, and these types of effectors might contribute to insect adaptation to host plants during coevolution.
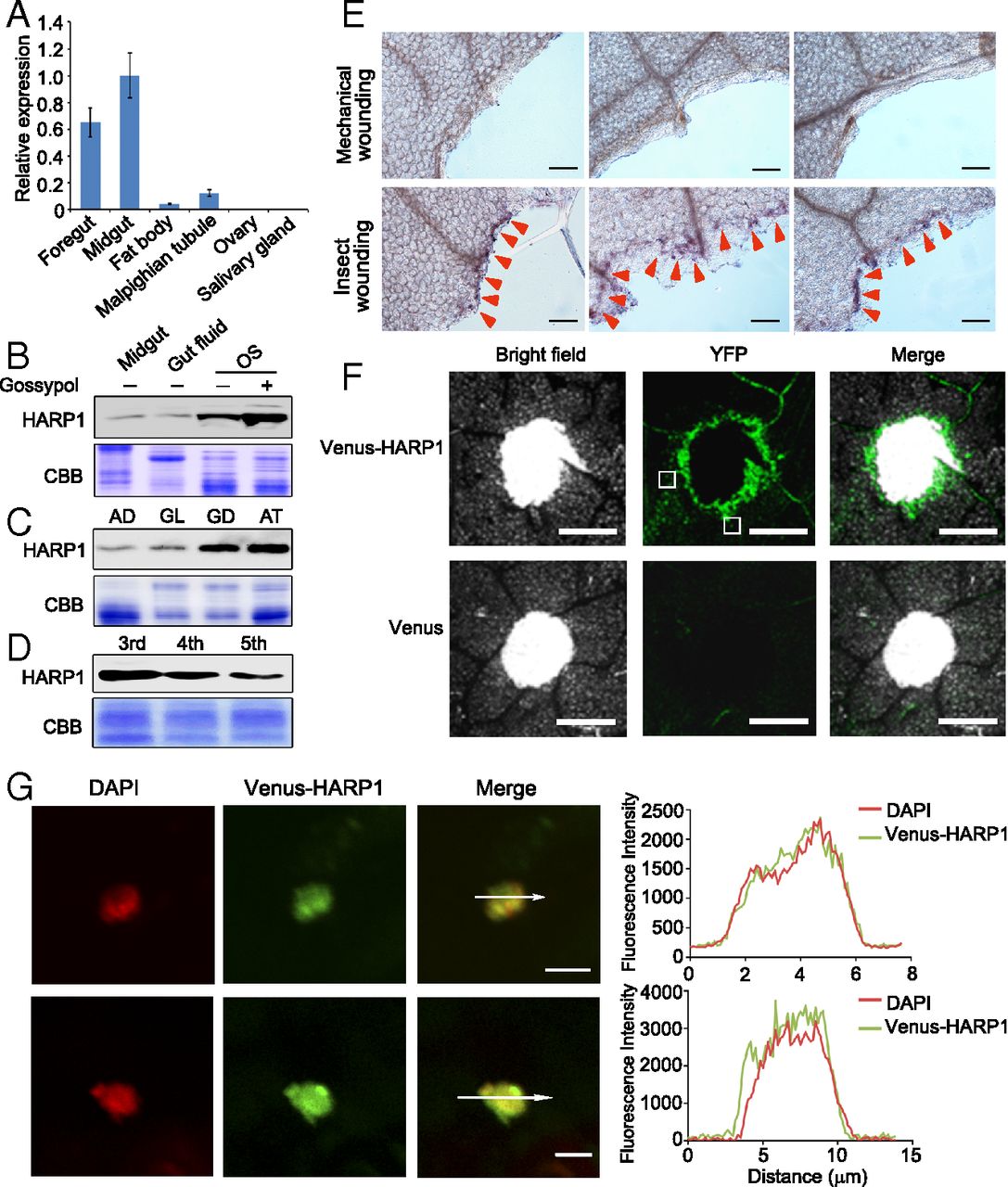
Figure 1. HARP1 in H. armigera OS migrates into the leaf cells through the wounding damage sites. (A) qRT-PCR analysis of HARP1 transcripts in indicated tissues of fifth-instar larvae. The expression level in midgut was set to 1. Error bars represent ± SD (n = 3 biological replicates). (B–D) Immunoblot detection of HARP1 protein level. The protein amount in each loading was quantified by Bradford assay and visualized by Coomassie Brilliant Blue (CBB) staining. All of the experiments were repeated at least two times, and the results were consistent. In B, the fourth instar larvae were fed on artificial diet supplemented with (+) or without (−) 0.1% gossypol for 1 d, and total proteins were collected from midgut, gut fluid and OS. In C, the OS was collected from the fourth instar larvae fed on artificial diet (AD), glandless (GL), or glanded (GD) cotton and A. thaliana (AT) leaves for 1 d. In D, the OS was collected from the indicated instar larvae that were fed on AT leaves for 1 d. (E) Whole amount immunohistochemistry detection of HARP1 at the chewing sites (red arrows) of Arabidopsis leaves. The mechanical wounding leaves were used as negative control. Anti-HARP1 antibody was used to detect HARP1 in B–E. (F and G) Translocation of the Venus-HARP1 fusion protein into plant cells through damage sites. The Arabidopsis leaves were punched and incubated with the protein solutions of Venus-HARP1 or Venus for 1 h and washed for three to four times to remove the extra proteins that adhered on the leaf surface. The boxes indicate the location as shown in G. (F) Venus-HARP1 but not Venus was detected at the wounded sites. (G) A portion of Venus-HARP1 was located in the nucleus of leaf cell. Fluorescence intensity in cross-section (white arrow) is shown. (Scale bars: E, 100 μm; F, 500 μm; G, 5 μm.)
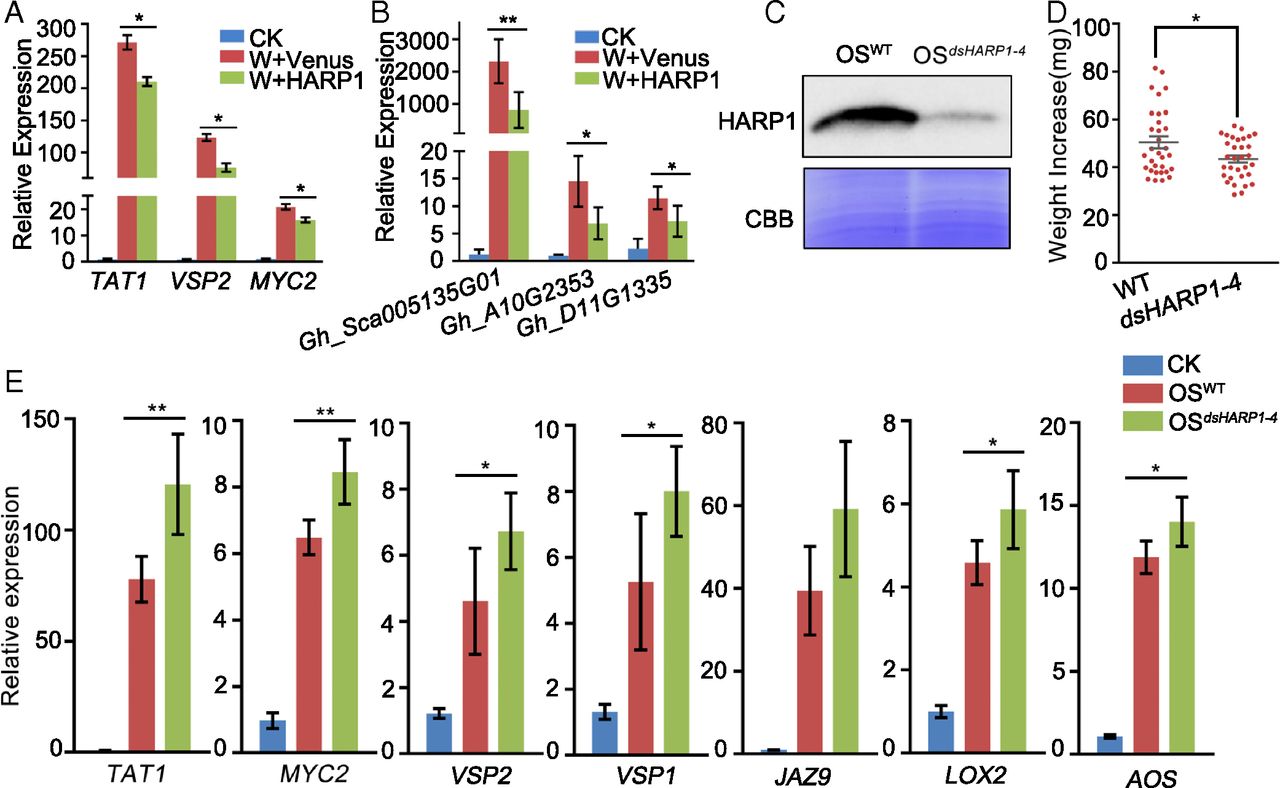
Figure 2. HARP1 reduces plant response to wounding. (A and B) Genes were less induced upon wounding in the presence of HARP1. Arabidopsis (A) and cotton (B) leaves were mechanically wounded and painted with the prokaryotically expressed HARP1 (W+HARP1) or Venus (W+Venus) solutions (1 mg/mL) on the wounded sites. Samples were collected 4 h later, and the gene expressions were detected by qRT-PCR. The expression in unwounded plants (CK) was set to 1. Data were analyzed by Student’s t test. *P < 0.05, **P < 0.01. Error bars represent ± SD (n = 3 biological replicates in A, n = 5 biological replicates in B). All of the experiments were repeated three times, and the results were consistent. (C–E) The impacts of HARP1 reduction in OS on the larval adaptation to plants. (C) HARP1 accumulation was reduced in OS from the larvae fed on 35S:dsHARP1 plants. Third-instar larvae were fed with wild-type (WT) and 35S:dsHARP1-4 plant leaves for 4 d. HARP1 level in OS samples from the larvae fed on the WT (OSWT) and the 35S:dsHARP1-4 (OSdsHARP1-4) Arabidopsis leaves were detected by immunoblot. The amount of total proteins in each loading was quantified by Bradford assay and visualized by CBB staining. (D) The growth of H. armigera larvae fed with 35S:dsHARP1-4 plants was inhibited compared with those fed with wild type. Weight increases of third-instar larvae fed on Arabidopsis leaves of WT and 35S: dsHARP1-4 (dsHARP1-4) for 4 d were recorded. Data were analyzed by Student’s t test. *P < 0.05. Error bars represent ± SEM (n = 32). (E) Gene expressions in plant leaves after the treatment with the OS samples as described in C. Arabidopsis leaves were wounded and painted with OSWT and OSdsHARP1-4, respectively; samples were collected 2 h after treatment. qRT-PCR was used to detect gene expressions. The gene expression in the unwounded plants (CK) was set to 1. *P < 0.05, **P < 0.01. Error bars represent ± SD (n = 5 biological replicates).
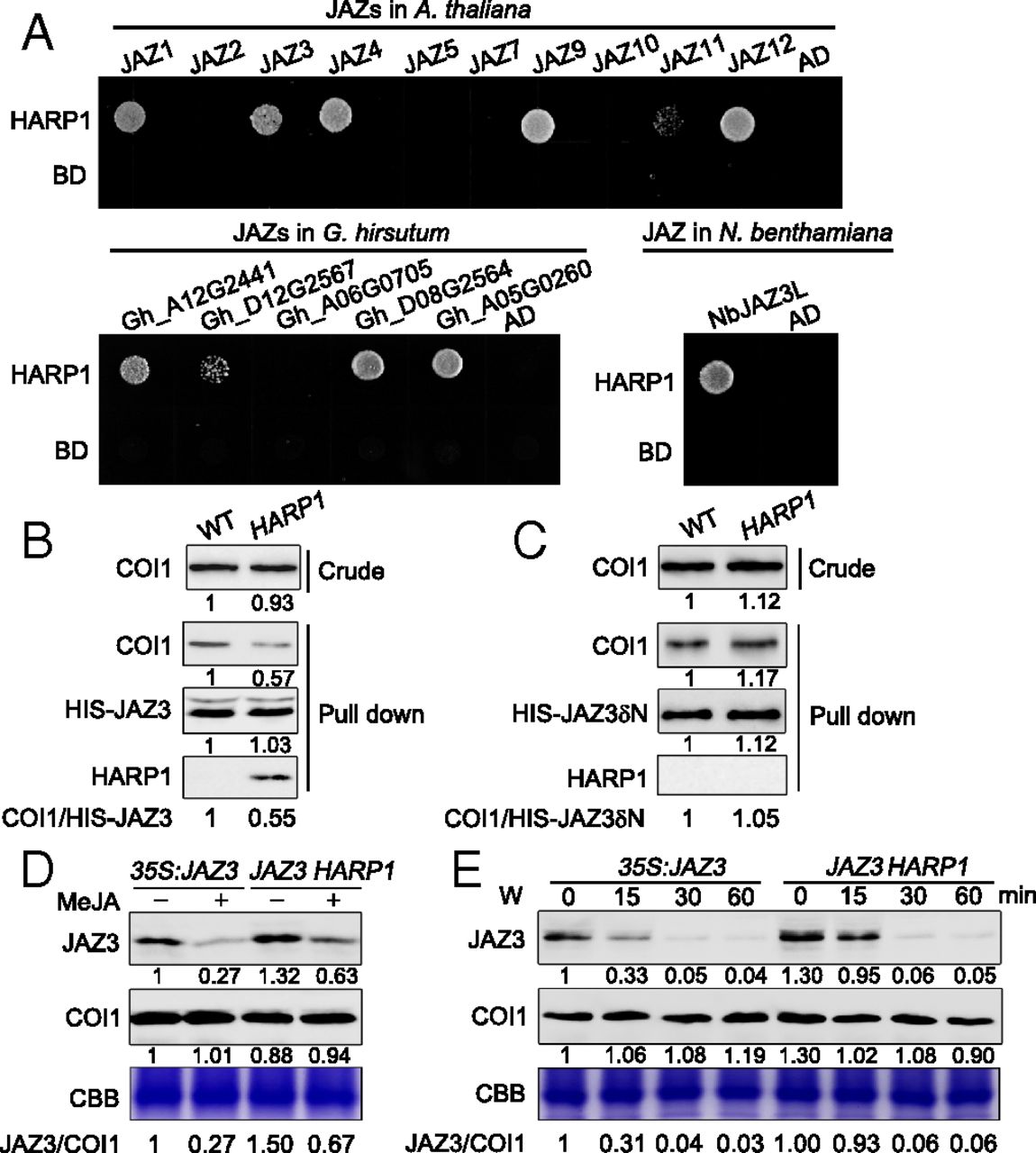
Figure 3. HARP1 interacts with and stabilizes JAZ proteins. (A) Yeast two-hybrid assay. HARP1 was fused to GAL4 DNA-binding domain (BD), JAZ proteins of Arabidopsis (A. thaliana), cotton (G. hirsutum), and tobacco (N. benthamiana) were fused to GAL4 activation domain (AD), respectively. Interactions were examined with 1 mM 3-amino-1,2,4-triazole. (B and C) HARP1 reduces COI1-JAZ3 coprecipitation. Recombinant proteins of HIS-JAZ3 (B) and HIS-JAZ3δN (C) were incubated with total leaf proteins of the wild-type (WT) and 35S:6MYC-HARP1-1 (HARP1) Arabidopsis in the presence of 50 μM Coronatine. Anti-COI1 antibody was used to detect COI1 level before (Crude) or after (Pull down) immunoprecipitation. Anti-MYC antibody was used to detect the 6MYC-HARP1 (HARP1), and Anti-HIS antibody was used to detect HIS-JAZ3 and HIS-JAZ3δN. Band intensity was quantified by ImageJ and was shown under each blot. The intensity of the WT was set to 1. The relative COI1/HIS-JAZ3 ratios were listed in the bottom. The coimmunoprecipitation of COI1 with HIS-JAZ3 but not HIS-JAZ3δN was inhibited in the presence of HARP1. (D and E) HARP1 increases JAZ3 accumulation. The JAZ3-HA level is more stable in 35S:JAZ3-HA 35S:6MYC-HARP1 than in 35S:JAZ3-HA after 50 μM MeJA (D) or wounding (E) treatment. The plant leaves were collected 45 min after MeJA treatment or at the indicated time after wounding. Anti-HA antibody was used to detect JAZ3-HA (JAZ3). Band intensity was quantified by ImageJ and was shown under each blot. The intensity of the untreated 35S:JAZ3 –HA sample was set to 1. The relative JAZ3/COI1 ratios were listed in the bottom. The amount of total proteins in each loading was quantified by Bradford assay and visualized by CBB staining.
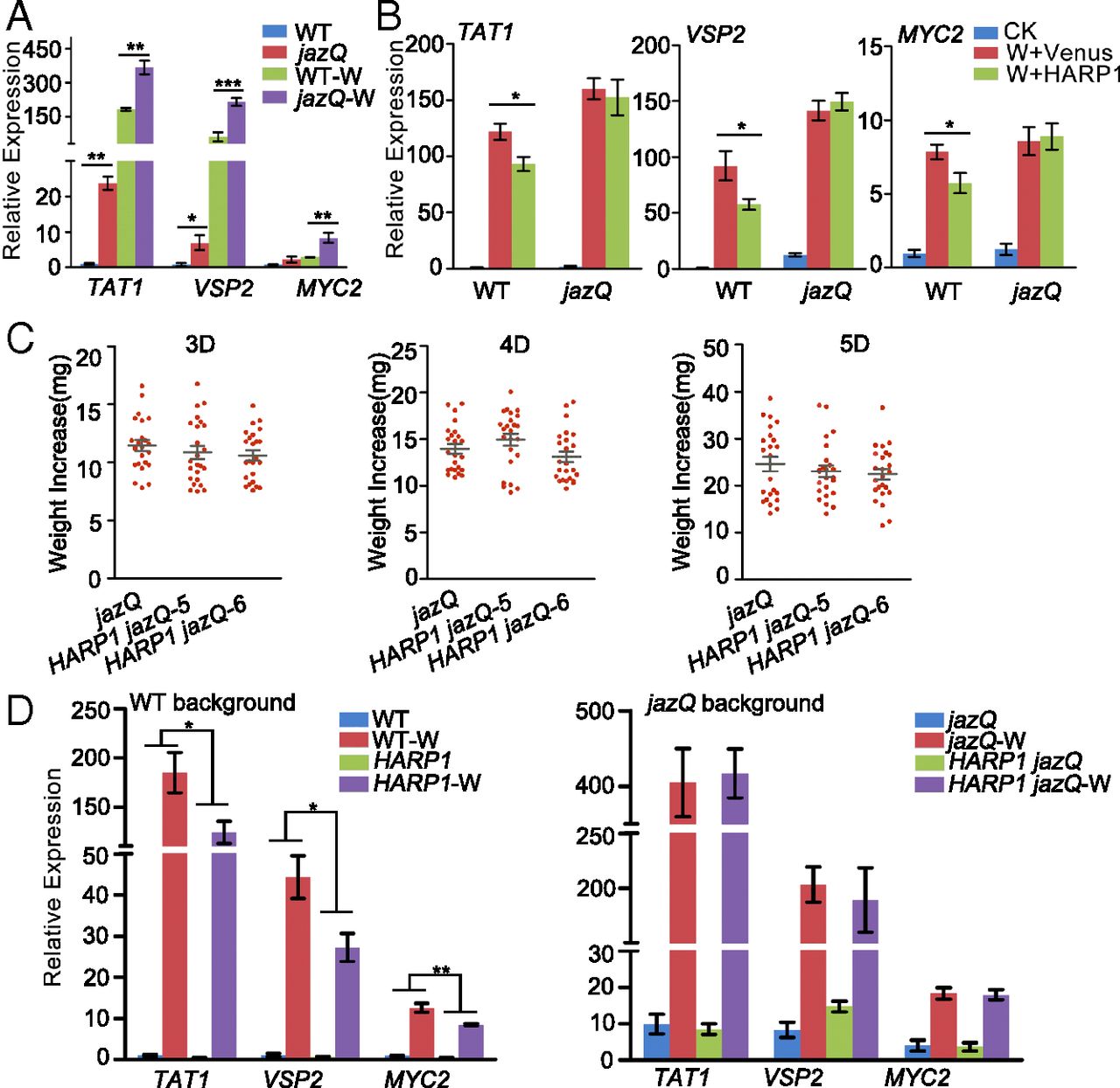
Figure 4. Wounding responses and insect resistance were not obviously affected by HARP1 in the jaz quintuple mutant jazQ. (A) Wounding responses were higher in jazQ. The wild-type (WT) and jazQ leaves were treated with mechanical wounding; gene expressions in leaves of the unwounded and the wounded (W) plants were detected by qRT-PCR 4 h after treatment. The gene expressions in unwounded WT were set to 1. Data were analyzed by Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001. Error bars represent ± SD (n = 3 biological replicates). (B) The gene inductions were not affected by HARP1 in the jazQ mutant upon wounding. The prokaryotically expressed HARP1 (W+HARP1) and Venus (W+Venus) was painted on the wounding sites of WT and jazQ plant leaves. The unwounded plants were used as control (CK). Gene expressions were detected by qRT-PCR 4 h after treatment. The gene expressions in unwounded WT were set to 1. Data were analyzed by Student’s t test. *P < 0.05. Error bars represent ± SD (n = 5 biological replicates). (C) H. armigera larvae gained similar weight increase when fed on 35S:6MYC-HARP1 jazQ (HARP1 jazQ) and jazQ leaves. The third-instar larvae were fed with plant leaves for indicated days, and the weight increases were measured (n = 24). (D) 35S:6MYC-HARP1 jazQ and jazQ plants exhibited similar gene inductions upon wounding. The HARP1 expressing plants under WT and jazQ background were treated with wounding (W) and samples of WT, 35S:6MYC-HARP1 (HARP1), jazQ, and 35S:6MYC-HARP1 jazQ (HARP1 jazQ) were collected 4 h after treatment. Gene expressions were detected by qRT-PCR. The expression in the unwounded WT was set to 1. Data were analyzed by two-way ANOVA followed by multiple comparisons (Tukey test). (*P < 0.05, **P < 0.01). Error bars represent ± SD (n = 5 biological replicates). All of the experiments were repeated at least two times, and the results were consistent.
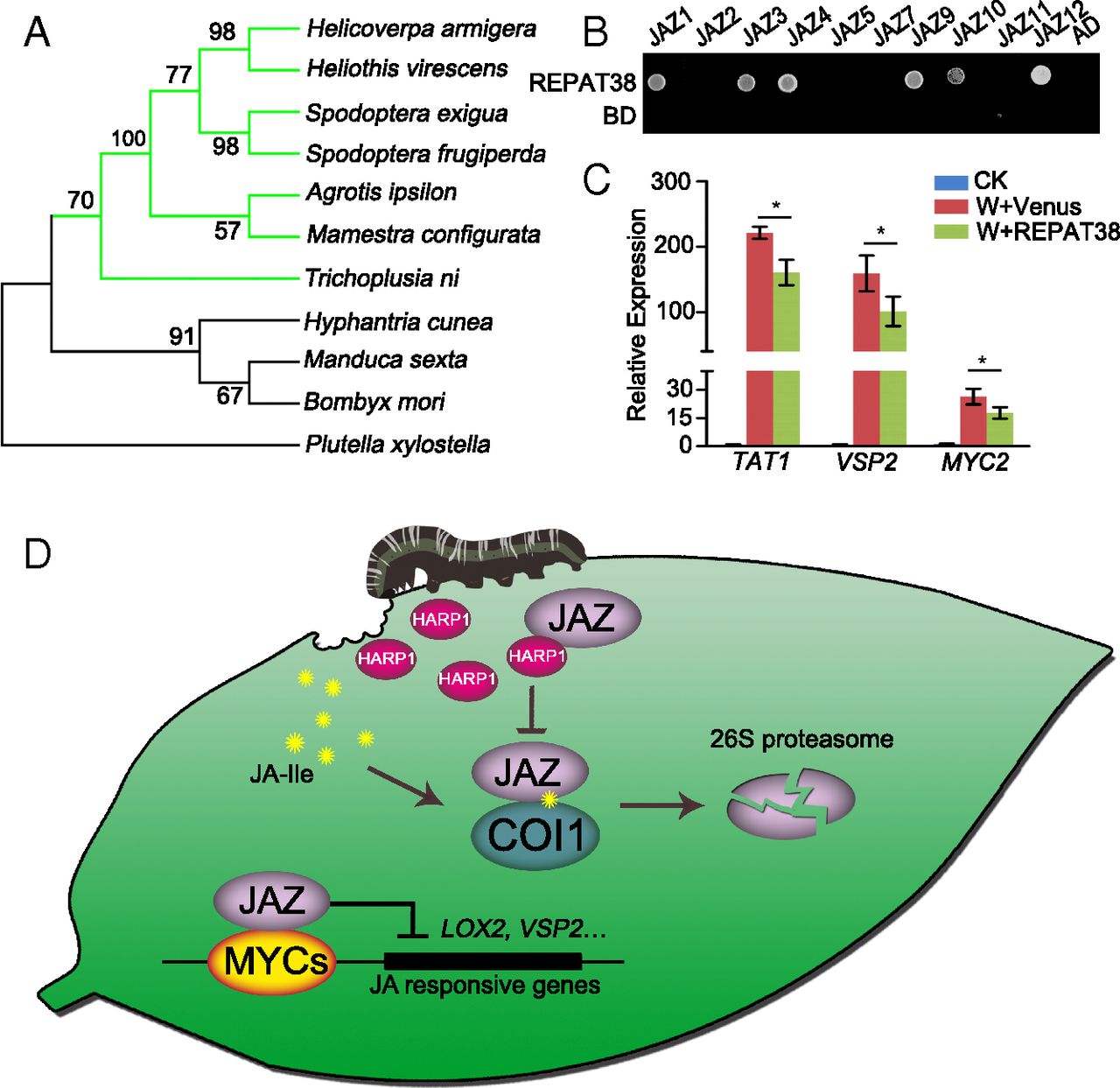
Figure 5. HARP1-like proteins highly conserved in Noctuidae and have similar function as effectors. (A) Phylogenetic analysis of HARP1 proteins in Lepidoptera insects. HARP1 (H. armigera) and the homologous proteins in Heliothis virescens, Spodoptera frugiperda, S. exigua, Agrotis ipsilon, Mamestra configurata, Trichoplusia ni, Hyphantria cunea, Manduca sexta, Bombyx mori, and P. xylostella were analyzed. The green lines represent these insects belongs to Noctuidae. (B) Yeast two-hybrid assay. REPAT38 was fused to GAL4 DNA-binding domain (BD), and the indicated JAZs were fused to GAL4 activation domain (AD), respectively. Interactions were examined with 1 mM 3-amino-1,2,4-triazole. (C) Gene expressions were less induced upon wounding in the presence of REPAT38. Samples were collected 2 h after treatment (W), and the indicated gene expressions were detected by qRT-PCR. The gene expression in unwounded plant was set to 1. Data were analyzed by Student’s t test. *P < 0.05, Error bars represent ± SD (n = 3 biological replicates). (D) Working model of HARP1 in plant–insect interaction. When H. armigera larvae feed on plant leaves, the wounding damage triggers the JA signaling. Meanwhile, the larval effector HARP1 in OS is released into plant cells and interacts with JAZs to prevent COI1–JAZ interaction and JAZ degradation, repressing the JA-regulated defense response.