Welcome to Mao lab!
Transcriptome analysis of three cotton pests reveals features of gene expressions in the mesophyll feeder Apolygus lucorum
Abstract The green mirid bug Apolygus lucorum is an agricultural pest that is known to cause damage to more than 150 plant species. Here, we report the transcriptomes of A. lucorum at three different developmental stages (the second and fifth instar nymphs and adults). A total of 98,236 unigenes with an average length of 1,335 nt was obtained, of which 50,640 were annotated, including those encoding digestive enzymes and cytochrome P450s. Comparisons with cotton bollworm and cotton aphid transcriptomes revealed distinct features of A. lucorum as a mesophyll feeder. The gene expression dynamics varied during development from young nymphs to adults. The high-quality transcriptome data and the gene expression dynamics reported here provide valuable data for a more comprehensive understanding of the physiology and development of mirid bugs, and for mining targets for their control.
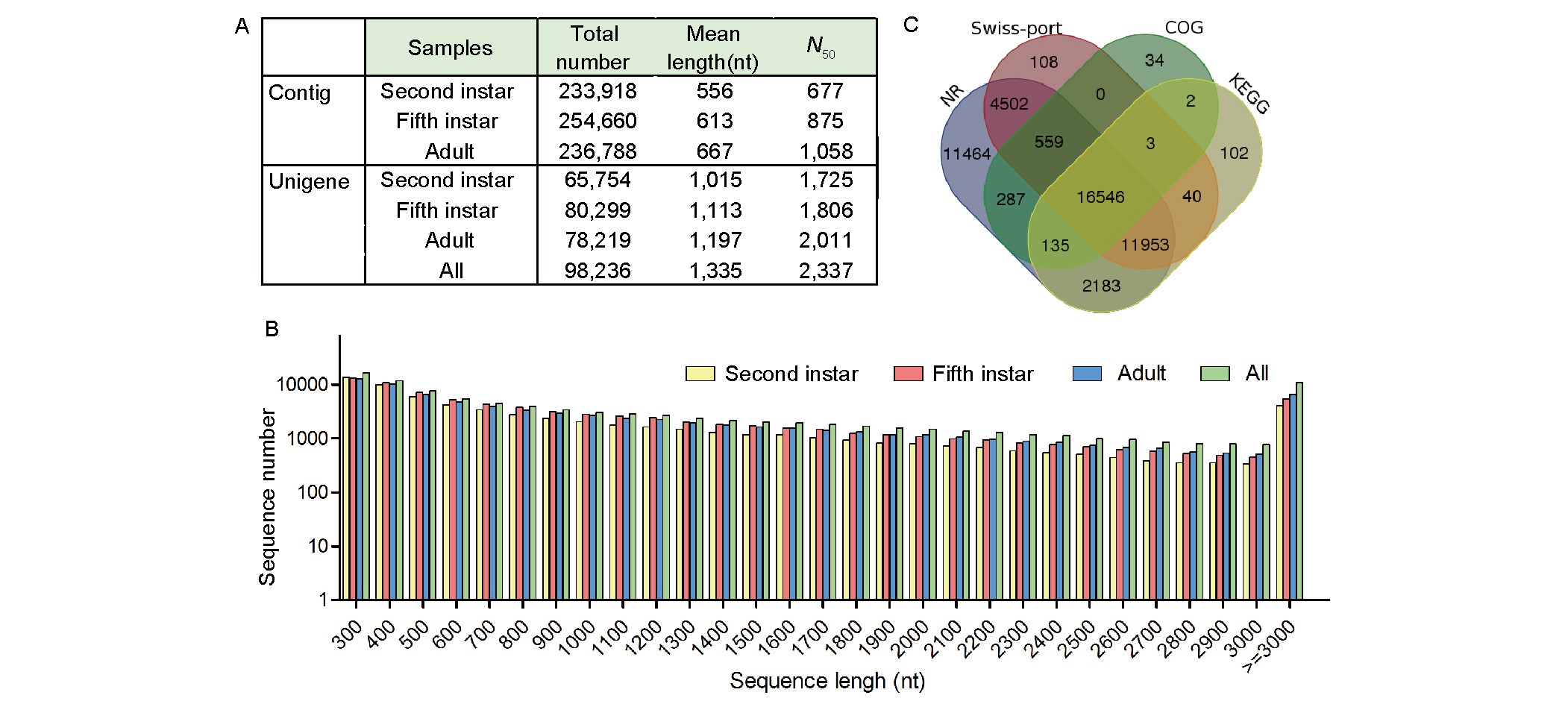
Figure 1. Unigenes of A. lucorum assembled from transcriptomes. A, Summary of assembly statistics of the transcriptomes at three developmental stages. B, Length distribution of unigenes assembled. C, Distribution of sequence similarity searching results illustrated by Venn diagrams. The numbers are the sum of transcripts annotated by NR, Swiss-prot, COG, and KEGG. Overlapping regions among the four circles contain the number of transcripts sharing BLASTx similarity with respective databases.
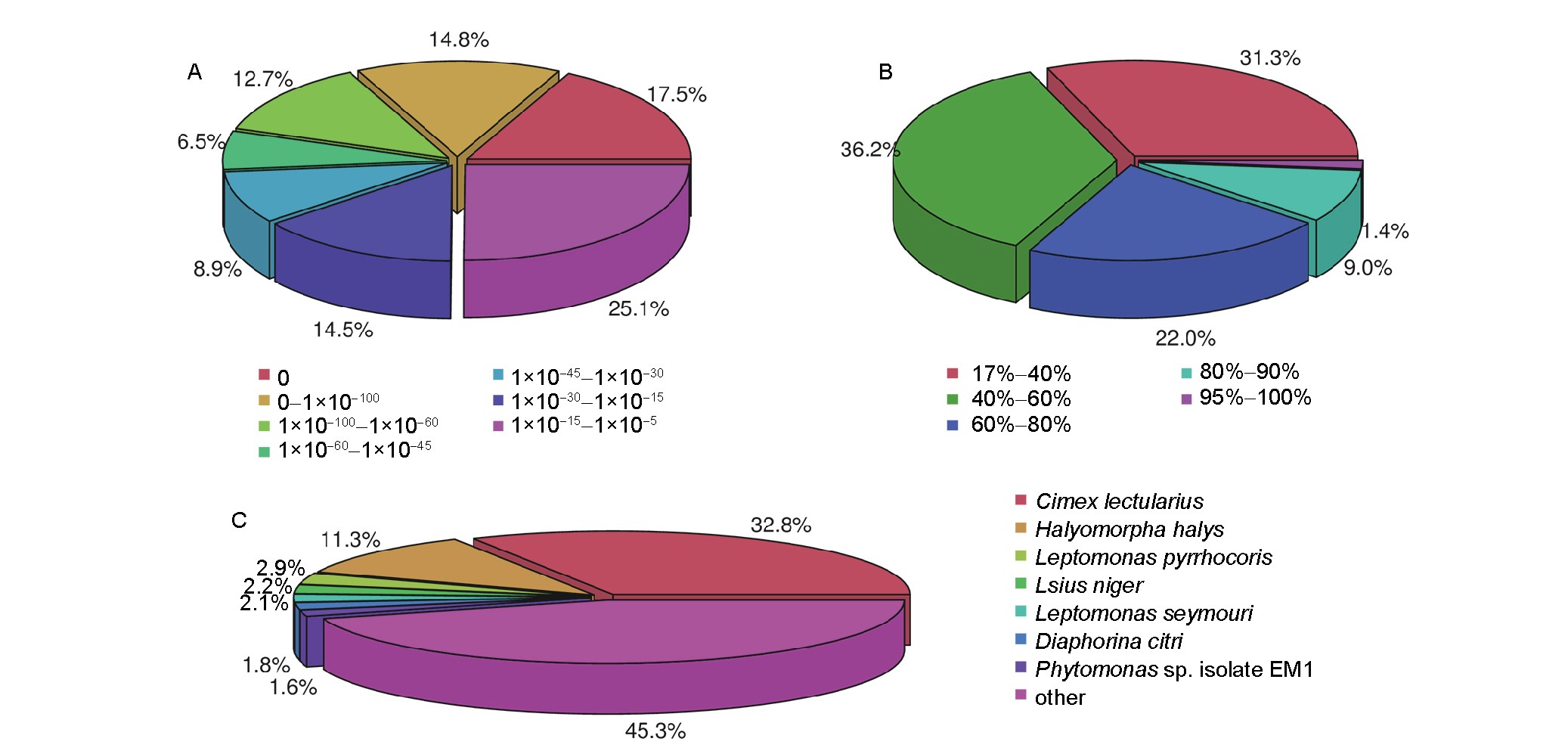
Figure 2. Characteristics of homology search. Distributions of BLASTx matches of A. lucorum unigenes against NR database with respect to E-values (A), gene identity (B), and insect species (C), with which the homologous genes were matched with.
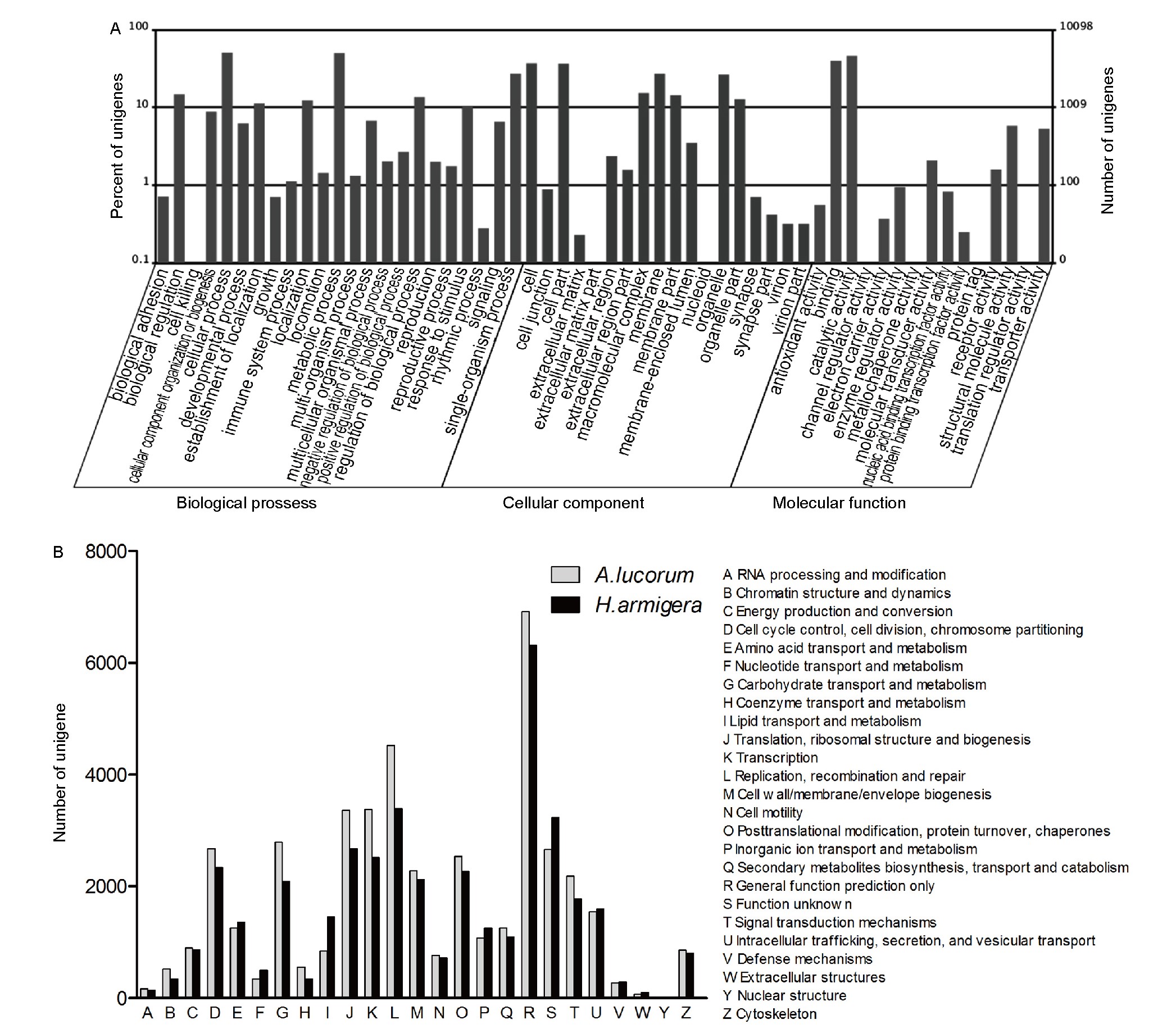
Figure 3. GO and COG annotations of unigenes. A, Categorization of A. lucorum unigenes with GO. B, Classification of A. lucorum and H. armigera unigenes with COG. Numbers of A. lucorum and H. armigera unigenes annotated into COG categories are shown.
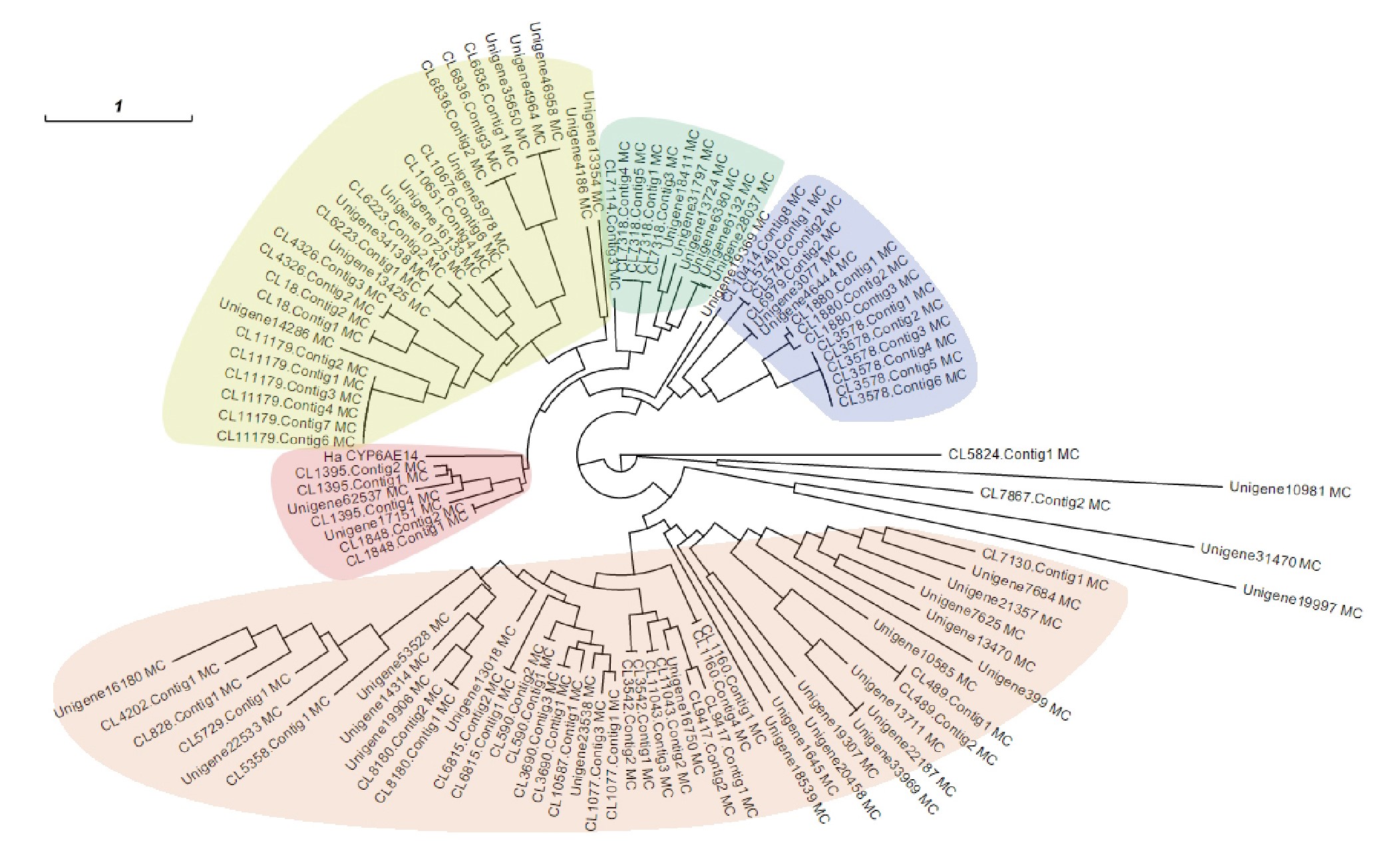
Figure 4. Phylogenetic tree of A. lucorum P450s. Deduced amino acid sequences of unigenes (>300 amino acid residues in length) were used to construct the phylogenetic tree. The five major clades are shaded in different colors, and a cotton bollworm P450 HaCYP6AE14 is included in the analysis and its location in the clade is shaded in pink together with seven A. lucorum P450s.
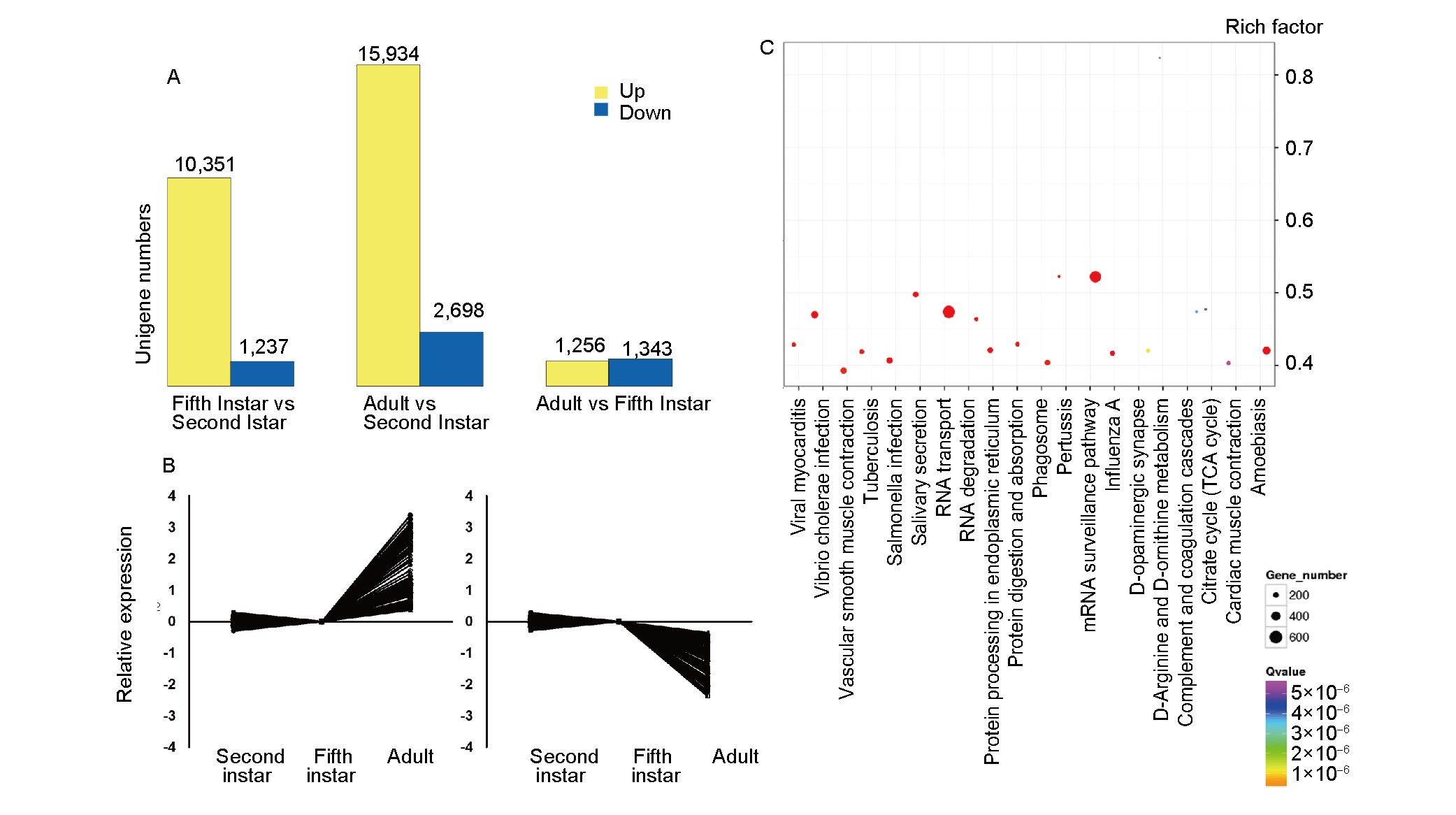
Figure 5. Differentially expressed genes (DEGs) of A. lucorum at three developmental stages. A, Numbers of up- and down-regulated unigenes at different developmental stages. Yellow bars represent up-regulated and blue bars stand for down-regulated unigenes. B, Expression patterns of unigenes up- and downregulated in adults. Unigenes were selected by FPKM analysis. The FPKM for each unigene was above five at least in one sample, and the up/down-regulated unigenes in the adults compared to their expression levels in the second and the fifth instar nymphs were chosen (fold change >2 or <0.5, FDR<0.001). Y-axis stands for gene expression levels relative to the second instar nymphs (Log10 FPKM). C, Top 20 enrichment pathways for the adult vs. the second instar nymphs. Y-axis stands for the rich factor of each item. The size and color of each blot stand for the gene number and Q-value, respectively.
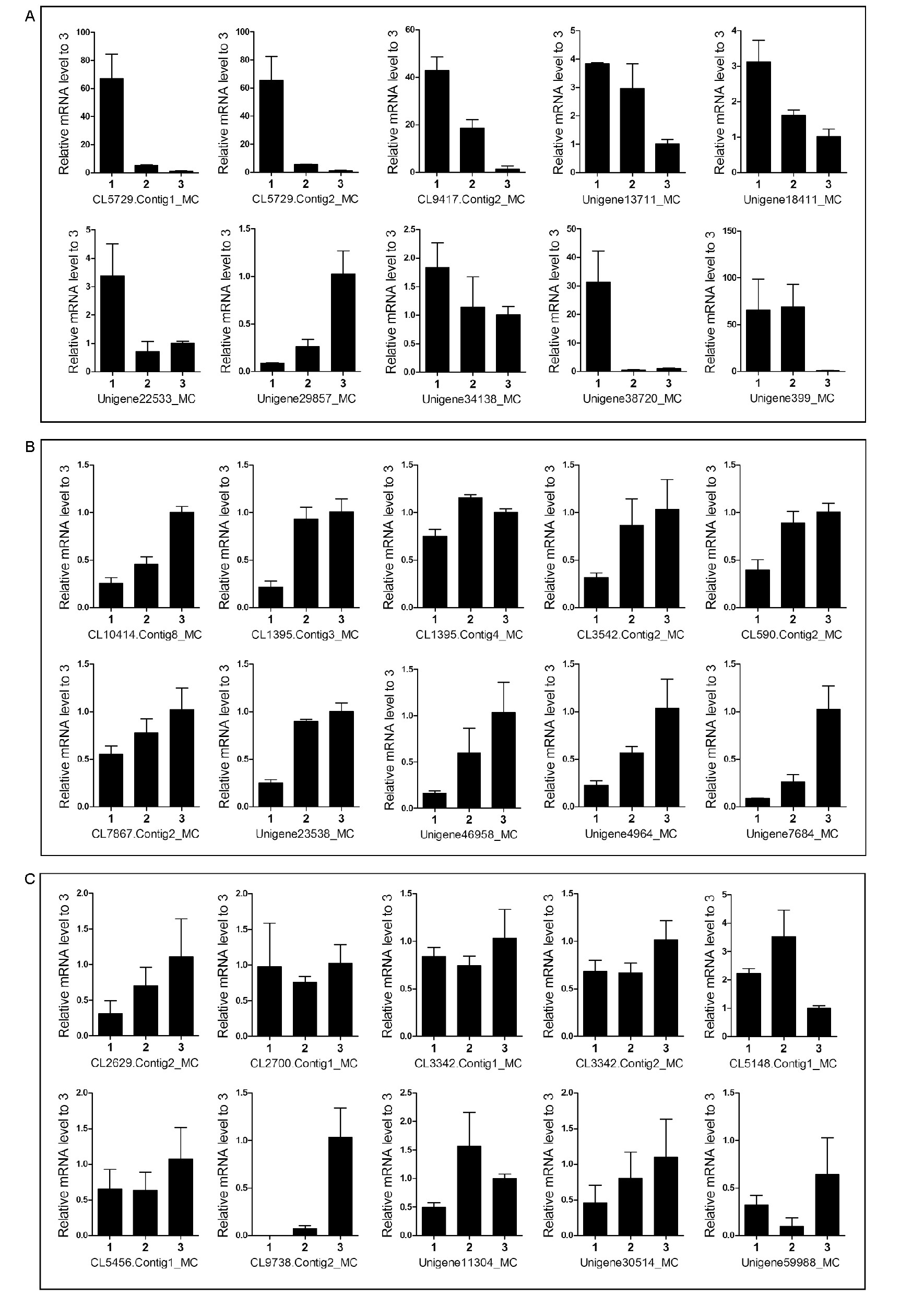
Figure 6. qRT-RCR analysis of 20 A. lucorum P450s and 10 trypsin-like proteases at three developmental stages. A, P450 genes down-regulated in adults. B, P450 genes up-regulated in adults. C, Trypsin-like protease genes. 1, the second instar nymphs; 2, the fifth instar nymphs; 3, the adults. The mRNA levels of the second and the fifth instar nymphs were normalized to adults and the relative mRNA levels are presented as means±SD (n=3).
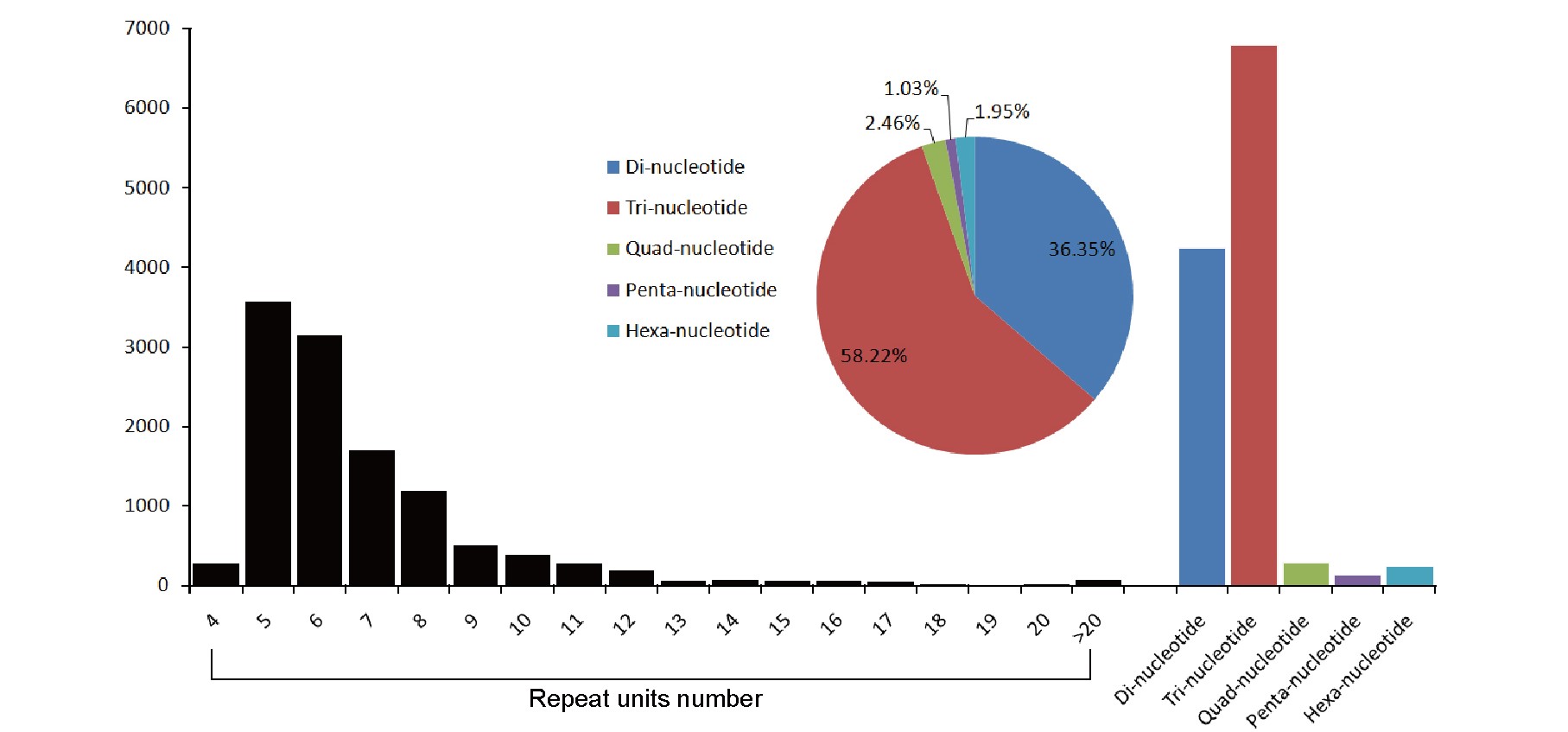
Figure 7. SSRs in A. lucorum transcriptomes. Black bars stand for the numbers of SSRs of different repeat units. Colored bars and pie chart stand for numbers and percentages of di-, tri-, quad-, penta-, and hexa-nucleotide repeats, respectively