
Recent research progresses
1. Design and construction of artificial biological systems for complex natural products biosynthesis
1.1 Strength prediction based rational design of fine-tuning regulatory elements
The design and construction of complex artificial biological systems or networks require a number of fine-tuning regulatory elements, e.g., promoters with different strengths. We have constructed complex non-linear artificial neural network (ANN) models to achieve higher predicting performance, and finally succeed in rational design of fine-tuning regulatory elements (Fig 1). The predicting methodology and designed sequences can be further applied for synthetic biology applications.
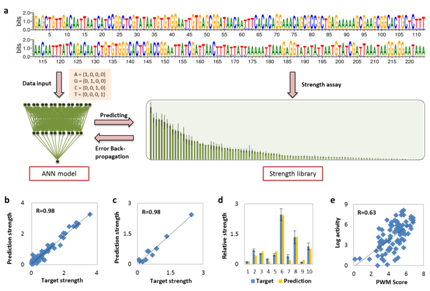
Fig 1. Strength prediction based rational design of fine-tuning regulatory elements. A, Workflow for regulatory element design using BP-ANN based method. B, Prediction accuracy for training dataset. C & D, Prediction accuracy for testing dataset. E, Prediction accuracy for a linear correlation method PWM.
1.2 Construction and application of genome-scale in silico metabolic models for strain improvement and network redesign
The genome-scale based in silico analysis platform was developed and used to direct the design and construction of microbial hosts for heterologous natural product production. Based on the flux balance analysis (FBA) method, we have developed a novel algorithm named as flux distribution comparison analysis (FDCA). The FDCA method was performed by separately calculating the flux distribution profiles for the wild-type objective (e.g., the maximum biomass formation rate) and expected objective (e.g., the maximum IPP synthesis rate) using flux balance analysis. Afterwards, the flux distribution landscapes of the two objectives were compared to identify the potential metabolic nodes (reactions) with significant differences. We further successfully used the FDCA method to optimize the production of 6-deoxy-erythronolide (6dEB) and isoprenoids in . coli.
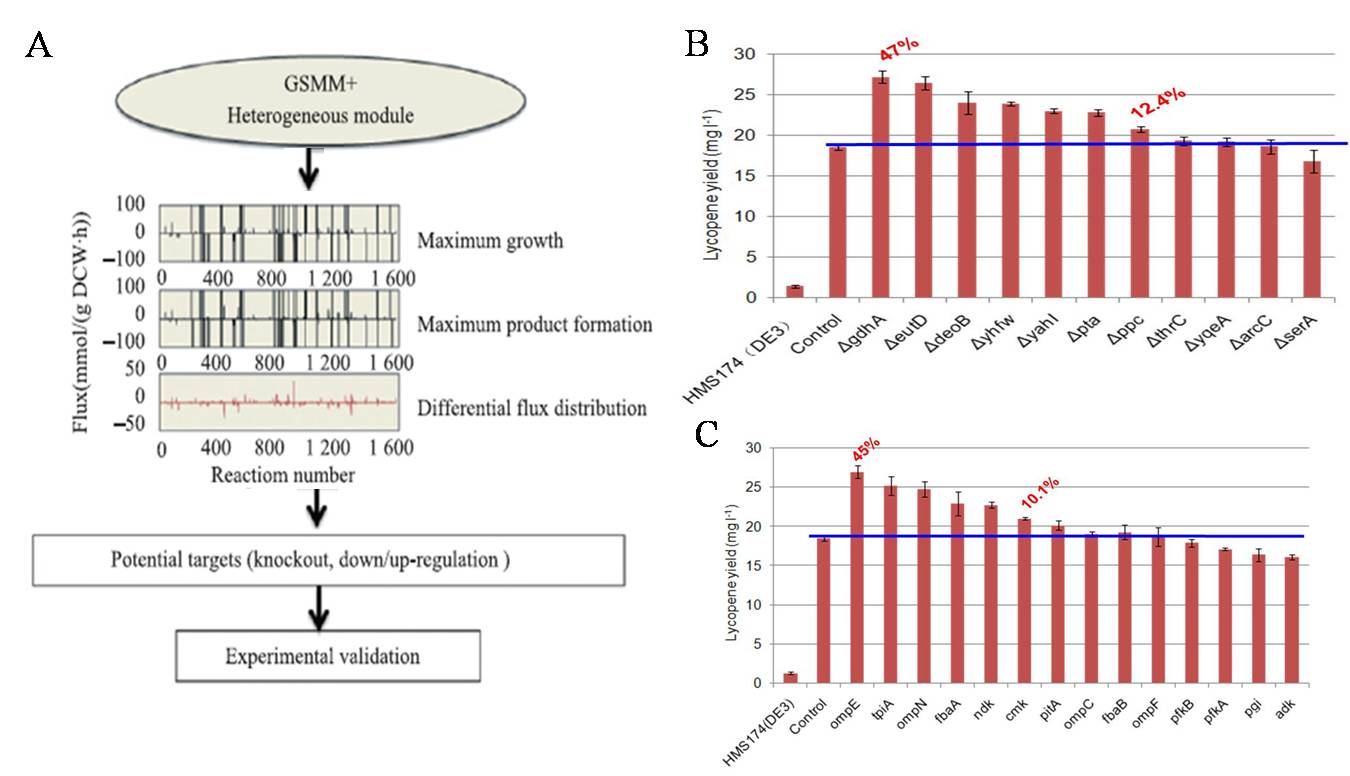
Fig 2. Prediction and identification of potential genetic targets for improving isoprenoids in E. coli. using FDCA method. A, The workflow of FDCA method. B & C, Experimental identification of potential deletion (B) and amplification targets (C) for improving lycopene production in E. coli.
1.3 Heterologous biosynthesis of important isoprenoids and DXP pathway engineering in E. coli
Our group has constructed a series of heterologous biosynthetic pathways in E. coli and successfully achieved the production of various important isoprenoids or their key intermediates in E. coli, including steviolglycosides (a highly natural sweetener), taxol (an anti-cancer drug), artemisinin (an anti-malaria drug) and lycopene.
The DXP pathway is theoretically proved to be superior to MVA pathway for IPPprovision. However, very little information about the control of DXP pathway has been reported. Consideringthe great diversities of natural DXP pathway genes on the control of transcriptional and enzymatic level, we have systematically expressedvarious bacteria (e.g., Bacillus, Streptomyces, Saccharopolyspora and Erwina) derived DXP pathway genes to improve isoprenoid productionin E. coli. (Fig 3). When co-expressing the two best modules dxs2 from S. avermitilis and idi from B. subtilis, the amorphadiene yield reached 6.1 g/L in a 5 L bioreactor. This is the highest reported yield when using DXP pathway for isoprenoid biosynthesis.
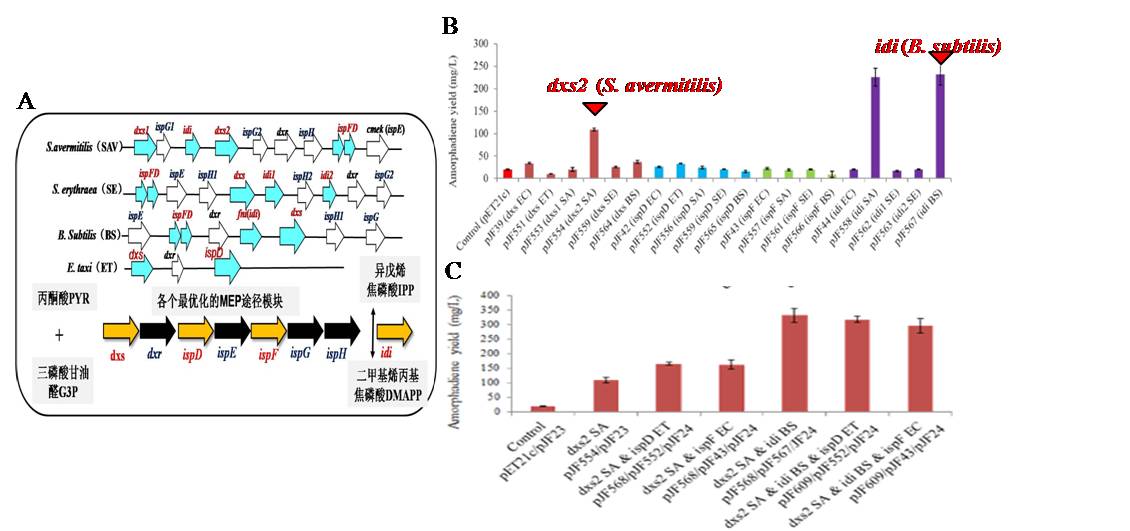
Fig 3. DXP pathway engineering in E. coli. using exogenous DXP pathway genes from different bacteria. A, Diversity of DXP pathway genes in bacteria. B, Amorphadiene production through over-expressing different exogenous DXP pathway genes. C, Amorphadiene production by combinatorial expression of different DXP pathway genes.
1.4 Engineering efflux pumps to improve biosynthesis of isoprenoids
Engineering isoprenoid efflux system to enhance the exportation of toxic heterologous products in microbial cells can release the adverse effects on cellular behaviors and further improve product yield. In our research, tripartite efflux pumps (e.g., AcrAB-TolC from E. coli.) with broad substrate spectrum have been successfully engineered to improve isoprenoid production in E. coli. (Fig 4).
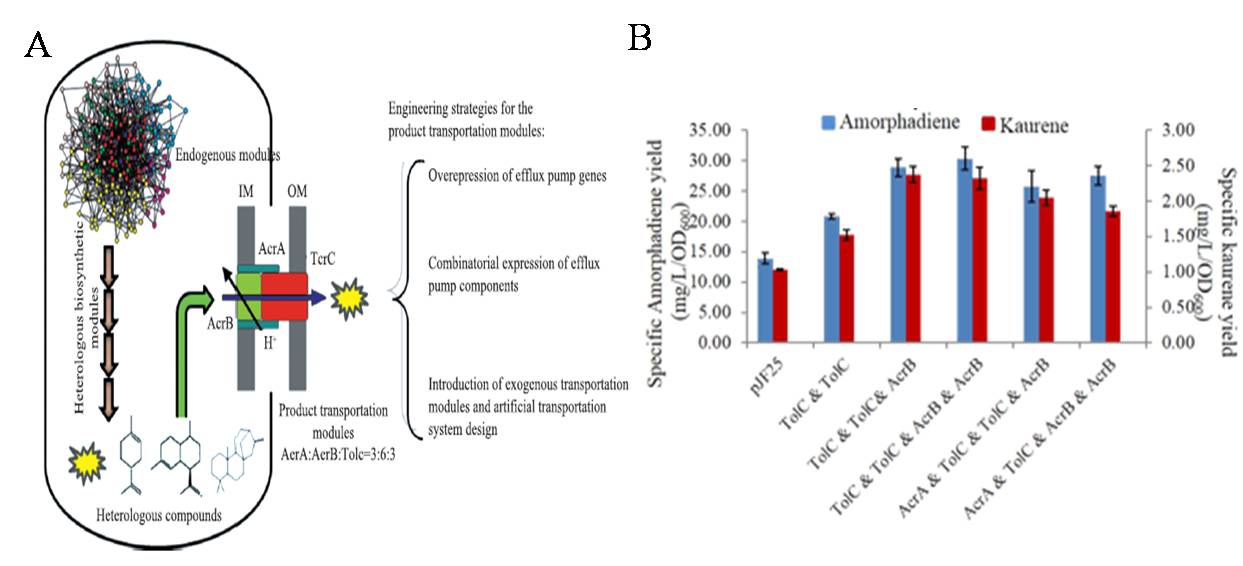
Fig 4. Engineering efflux pumps to improve biosynthesis of isoprenoids. A, Strategies for efflux pump engineering. B, Specific yields of amorphadiene and kaurene in efflux pump engineered strains. Empty plasmid pJF25 represents control.
2. Isolation of different gene/module/pathway resources for natural products biosynthesisfrom marine sedimentsand medicinal plants
The actinomycete inhabited in the marine sediments of the Yellow Sea and endophytic fungi from taxus x madiawere systematically mined by multiple approaches (e.g., morphological and molecular identification, bioactivities assays, and PCR screening). For example, 614 bacteria were recovered in the marine sediments of the Yellow Sea. 143 isolates displayed antibacterial activity. In addition, 5 isolates showed good anticancer activities. PCR screening indicated that 6 types of PKS-I, PKS-II, NRPS, isoprenoids, enediyne PKS, and halogenase genes were recovered in 58.14%, 51.16%, 30.23%, 46.51%, 30.23%, and 32.56% of 43 representative strains, respectively. For endophytic fungi, three representative species of the distinct genera (Guignardia, Fusarium, and Colletotrichum) capable of producing taxol were obtained. Among these 3 taxol-producing fungi, the highest yield of taxol was 720 ng/L by Guignardia mangiferae HAA-11. Genes coding taxadiene synthase (TS), 10-deacetylbaccatin III-10-O-acetyl transferase (DBAT), and C-13 phenylpropanoid side chain-CoA acyltransferase (BAPT) in the taxol biosynthetic pathway, were investigated in taxol-producing endophytic fungi (Fig 5). The lower similarities of ts and bapt between microbial and plant origin, indicating that fungal taxol biosynthetic cluster might be repeatedly invented during evolution, nor horizontal gene transfer from taxus species.
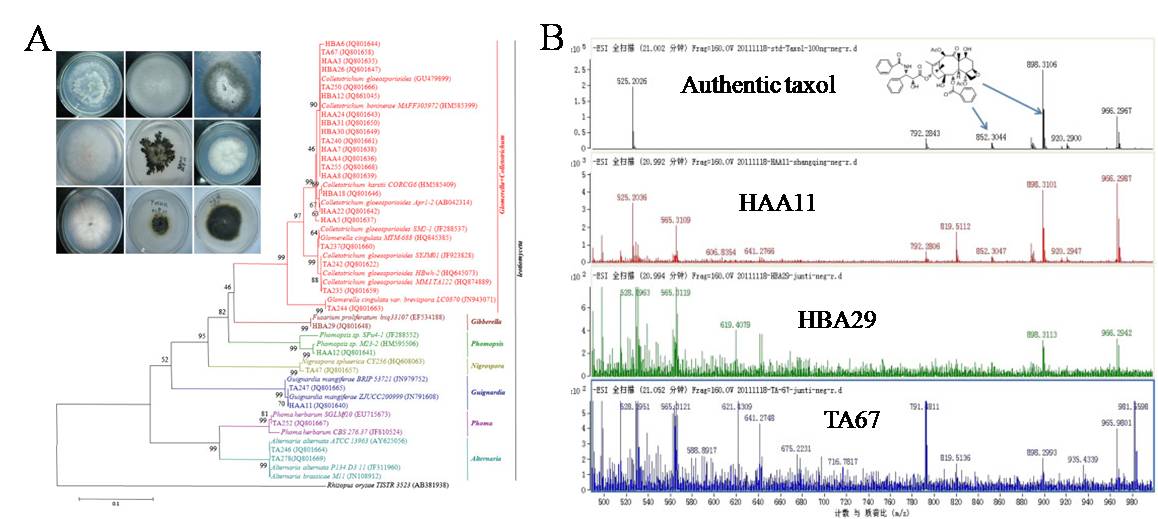
Figure 5. Diversity of endophytic fungi from taxus x madia (A) and production of taxol by endophytic fungi (B)


