Establishment of the mycorrhizal interaction in legumes involves a signalling pathway that also functions in the rhizobial symbiosis that leads to nodulation. The important question is how plant responses to rhizobial and mycorrhizal fungal infection use the same signalling pathway. Although several isolated genes function in the common signalling pathway has been identified using forward genetic screens, there are still few gaps in the signalling transduction from signal perception at plasma membrane to gene activation in nucleus. We propose that some of common signalling pathway genes are redundant in legumes, which may not be the case in other plant model systems. Given that the mycorrhizal symbiosis is conserved in legumes and rice, some of these genes, which are critical for mycorrhizal symbiosis in rice, are supposed to be important for mycorrhizal and rhizobial symbiosis in legumes. A genetic approach can be taken for isolating rice mutants that are defective in the mycorrhizal symbiosis, then Medicago can be used to study roles of these genes in rhizobial symbiosis.
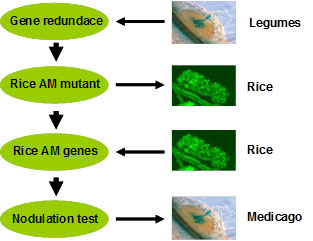
Work Pipline for nodulation research
Taking this approach, we propose to isolate one kind of AM defective mutant OsF, encoding OsF protein which interacts with OsRAM1, which is important for mycorrhizal colonization. In Medicago truncatula, we proposed OsF is also required for nodulation symbiosis. We would continue to underpin the role of OsF Medicago homology in nodulation and mycorrhizal signalling pathway and isolate more common signalling pathway genes using this genetic approach combined with bioinformatics and proteomics in future.
